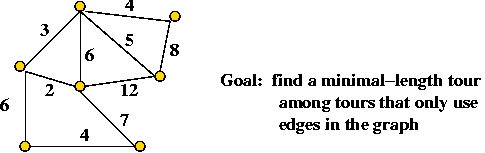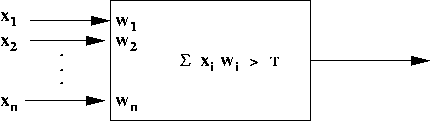Informal definition:
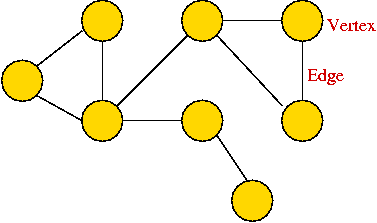
- A graph is a mathematical abstraction used to represent
"connectivity information".
- A graph consists of vertices and edges that
connect them, e.g.,
- It shouldn't be confused with the "bar-chart" or "curve" type
of graph.
Formally:
- A graph G = (V, E) is:
- a set of vertices V
- and a set of edges E = { (u, v): u and v are vertices }.
- Two types of graphs:
- Undirected graphs: the edges have no direction.
- Directed graphs: the edges have direction.
- Example: undirected graph
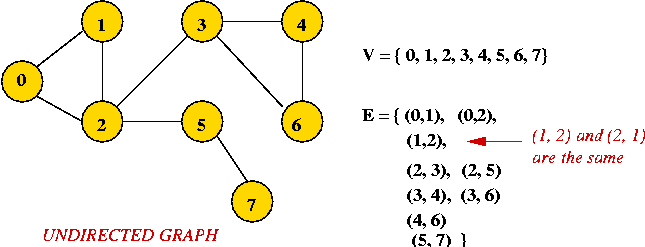
- Edges have no direction.
- If an edge connects vertices 1 and 2, either
convention can be used:
- No duplication: only one of (1, 2) or (2, 1)
is allowed in E.
- Full duplication: both (1, 2) and (2, 1)
should be in E.
- Example: directed graph
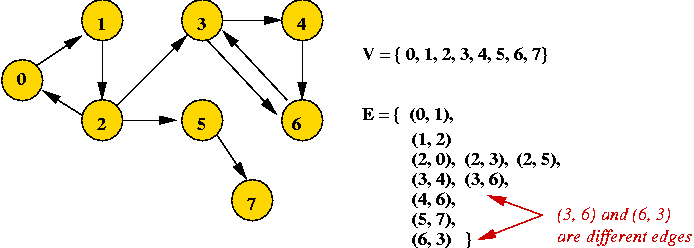
- Edges have direction (shown by arrows).
- The edge (3, 6) is not the same as the edge (6,
3) (both exist above).
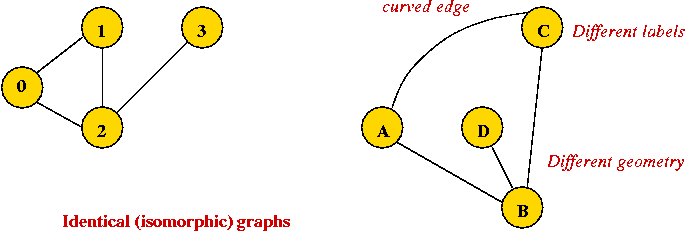
Depicting a graph:
- The picture with circles (vertices) and lines (edges) is
only a depiction
=> a graph is purely a mathematical abstraction.
- Vertex labels:
- Can use letters, numbers or anything else.
- Convention: use integers starting from 0.
=> useful in programming, e.g. degree[i] = degree
of vertex i.
- Edges can be drawn "straight" or "curved".
- The geometry of drawing has no particular meaning:
Exercise:
What is the maximum number of edges in an undirected graph with n vertices?
What is this number in order-notation?
Paths:
Why are graphs important?
- History:
- Applications:
- Fundamental mathematical construct to represent "connectivity".
- Appears in thousands of problems.
- Source of many classic problems: traveling salesman, routing,
spanning trees.
- Many "graph-structured" applications: networks,
transportation-systems, electronic circuits, molecules.
- Applications in Biology:
- Phylogenetic trees.
- Pathway databases.
- Genetic networks.
- Source of theory:
- Many important algorithms.
- Key to understanding algorithm design and analysis.
- Simple to describe, yet perplexing:
- Euler tour: easy problem.
- Hamiltonian tour: hard problem.
Optimization Problems
What are they?
- An optimization problem has "input" and a "measure"
=> different solutions possible, each of different value
=> Goal: find the "best" solution.
- The "measure" is sometimes called an objective function.
- Some problems have constraints: not all potentials solutions
are "acceptable".
Example: the Travelling Salesman Problem (TSP)
- Input: a collection of points (representing cities).
- Goal: find a tour of minimal length.
Length of tour = sum of inter-point distances along tour
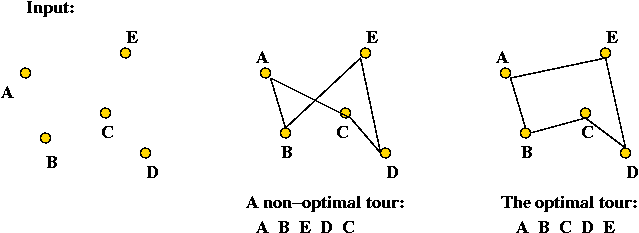
- Details:
- Input will be a list of n points, e.g., (x0, y0),
(x1, y1), ..., (xn-1, yn-1).
- Solution space: all possible tours.
- "Cost" of a tour: total length of tour.
=> sum of distances between points along tour
- Goal: find the tour with minimal cost (length).
- Note: strictly speaking, we have defined the Euclidean TSP.
=> There is also a graph version that we will not consider
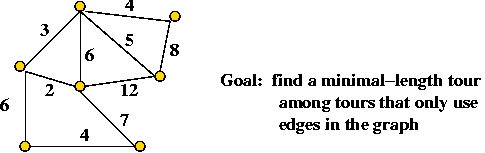
- Applications:
- Logistics: trucking and delivery problems.
- Machine operation (e.g., drilling a collection of holes)
Exercise:
For an n-point TSP problem, what is the size of the solution space
(i.e., how many possible tours are there)?
Example: the Bin Packing Problem (BPP)
- Input: a collection of items and unlimited bins.
- Goal: pack the items into as few bins as possible.
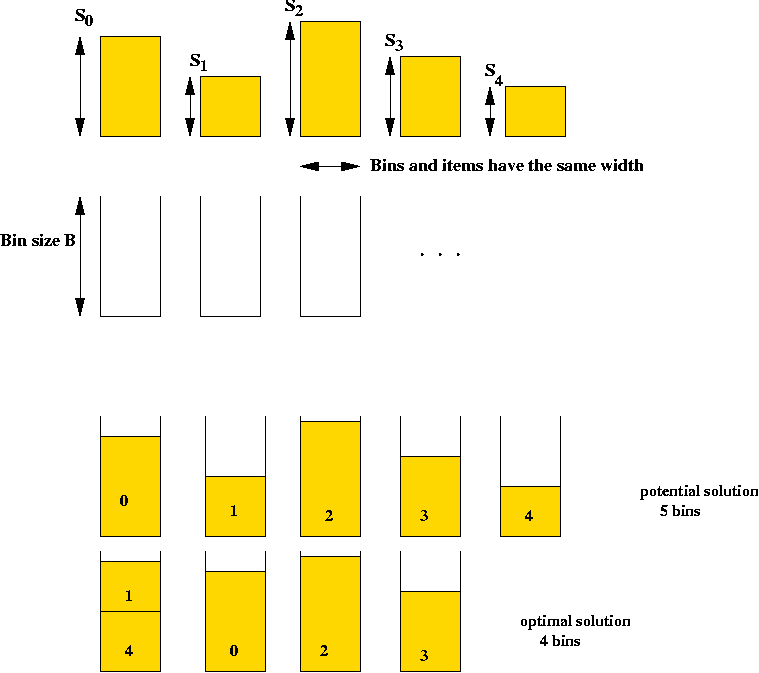
- Details:
- Input is a list of n integer bin sizes s0,
s1, ..., sn-1 and an integer bin size B.
- Solution space: all feasible packings
=> all assignments of items to bins such that no bin
overflows. (A constraint).
=> sum of sizes of items assigned to each bin is at most B.
- Goal: find the assignment that uses the fewest bins.
- Note: assume si < B.
- Alternate description:
- Item sizes: s0, s1, ...,
sn-1
where si < B.
- Define the assignment function:
| dij |
= |
1,
0, |
if item i is placed in bin j
otherwise |
- B = bin size.
- Goal: minimize k, the number of bins
such that:
- For each j,
s0dij + ... + sn-1dij < B
(all items assigned to bin j fit into the bin)
- For each i, dij + ... +
dij = 1
(each item is assigned to a bin)
Exercise:
Consider the following Bin Packing problem: there are three items with sizes
1, 2 and 3 respectively, and a bin size of 6.
Enumerate all possible assignments.
Example: Quadratic Programming Problem
- Input: n coefficients a0, a1,
..., an-1.
- Goal: minimize a0x02 +
a1x12 + ...
+ an-1xn-12.
- Constraint: x0 + ... + xn-1 = K
- Here, each xi is real-valued.
- Example:
Minimize 3 x02 + 4 x12
Such that x0 + x1 = 10
Types of optimization problems:
- The Quadratic Programming problem is very different
from both TSP and BPP.
- TSP and BPP are similar is some structural respects.
- Two fundamental types of problems:
- Discrete optimization problems: finite or countable solution space.
=> usually finite number of potential solutions.
- Continuous optimization problems: uncountable solution space
=> variables are real-valued.
- Examples:
- Discrete: TSP, BPP
- Continuous: Quadratic Programming problem.
- We will focus on discrete optimization problems.
Problem Size and Execution Time
Problem size:
- Typically, the "size" of a problem is the space required to
specify the input:
- Example: TSP
=> For an n-point problem, space required is: O(n).
- Example: BPP
=> For an n-item BPP, O(n) space is required.
- Non-Euclidean TSP
- Recall: a graph is given
=> required to use only edges in the graph.
- A graph with n vertices can have
O(n2) edge
=> O(n2) space required for input.
- Consider this TSP example:
- Input: "The points are n equally spaced points on the
circumference of the unit circle".
- How much space is required for an input of 10-points? For 1000-points?
=> O(1) !
- The above example is not general
=> we need O(n) space (worst-case) to describe
any TSP problem.
- Terminology: instance
- A problem instance is a particular problem (with the
data particular to that problem).
- Example: TSP on the 4 points (0, 1), (0.5, 0.6),
(2.5, 3.7) and (1, 4).
=> an instance of size 4.
- Another instance: TSP on the 5 points (0, 1), (0.5, 0.6),
(2.5, 3.7), (1, 4) and (6.8, 9.1).
=> an instance of size 5.
Execution time of an algorithm:
- What we expect of an algorithm:
- An algorithm is given its input and then executed.
- The algorithm should produce a candidate solution or report
that no solutions are possible.
- Example: TSP
- An algorithm is given the set of input points.
- The algorithm should output a tour after execution.
- Example: BPP
- An algorithm is given the item sizes and bin size as input.
- The algorithm should output an assignment
(Or report that some items are too large to fit into a bin).
- Output size:
- Example: TSP
- Output is a tour
=> O(n) output for n points.
- Example: BPP
- Output is an assignment.
- If assignment is specified as the function
| dij |
= |
1,
0, |
if item i is placed in bin j
otherwise |
then output could be as large as O(n2).
- Execution time:
=> Total execution time includes processing input and writing output.
Consider these two algorithms for TSP:
- Algorithm 1:
- Initially the set P = {0, ..., n-1 } and the set
Q is empty.
- Move 0 from P to Q.
- Repeat the following until P is empty:
- Suppose k was the point most recently added to
Q.
- Find the point in P closest to k and move
that to Q.
- Output points in the order in which they were added to Q.
- Algorithm 2:
- Generate a list of all possible tours and place in an array
(of tours).
- Scan array and evaluate the length of each tour.
- Output the minimum-length tour.
Exercise:
Consider the following 4 input points: (0,0), (1,0), (1,1) and
(0,-2).
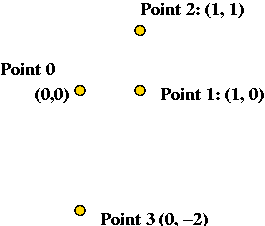
- Show the steps in executing each algorithm on this input.
- What is the complexity (execution time) of Algorithm 1 on an
input of size n?
- What is the complexity of Algorithm 2 on an
input of size n?
Polynomial vs. exponential complexity:
- An algorithm has polynomial complexity or runs in
polynomial time if its execution time can be bounded by a polynomial function
of its input size.
- Example: An algorithm takes O(n3) (worst-case) on input of size
n
=> algorithm is a polynomial-time algorithm.
- Requirements for the polynomial:
- Highest power in the polynomial should be a constant (i.e.,
not dependent on n.
- Polynomial should be finite (not an infinite sum of terms).
- Algorithms that run slower-than-polynomial are said to have
exponential complexity:
- Typically, the (worst-case) running time is something like
O(an).
- Note: factorials have (like O(n!)) have exponential
complexity
=> From Stirling's formula: n! = O(sqrt(n) * (cn)n).
Exercise:
Which of these are polynomial-time algorithms:
- Algorithm 0 runs in time O( (n2 + 3)4 ).
- Algorithm 1 runs in time O(n log(n)).
- Algorithm 2 runs in time O(nn).
- Algorithm 3 runs in time O(nlog(n)).
- Algorithm 4 runs in time O( (log n)3 ).
Summary:
- Many important optimization problems are
discrete optimization problems.
- For these problems:
- It's easy to find simple algorithms of polynomial complexity
=> but that are not guaranteed to find optimal solutions.
- The only algorithms that guarantee optimality take
exponential time.
Combinatorial Optimization Problems
A combinatorial optimization problem is:
- A set of states or candidate solutions S = {
s0, s1, ..., sm} .
- A cost function C defined on the states
=> C(s) = cost of state s.
- Goal: find the state with the least cost.
Example: TSP
- Each instance of TSP is a combinatorial optimization problem.
- Example: the 4-point TSP problem with points (0,1), (1,0),
(2,3) and (3,5)
- Does this have a set of "states" or "candidate solutions"?
=> Yes: S = { all possible tours }
= { [0 1 2 3], [0 1 3 2], [0 2 1 3], [0 2 3 1], [0 3 1
2], [0 3 2 1] }
- Is there a well-defined cost function on the states?
=> Yes: C(s) = length of tour s
e.g., C([0 1 2 3]) = dist(0,1) + dist(1,2) + dist(2,3) + dist(3,0).
- Is the goal to find the least-cost state?
=> Yes: find the tour with minimal length.
Example: BPP
- States: all possible assignments of items to bins.
- Cost function: C(s) = number of bins used in state s.
- Goal: find the state that uses the least bins (minimal-cost state).
Size of a combinatorial optimization problem:
- The input is usually of size O(n) or O(n2).
- TSP: list of n points.
- BPP: n item sizes and one bin size.
- Graph-based TSP: n vertices and up to
O(n2) edges.
- MST: n vertices and up to
O(n2) edges.
- The state-space is usually exponential in size:
- TSP: all possible tours.
- BPP: all possible assignments of items to bins.
- MST: all possible spanning trees.
- The output is usually of size O(n) or
O(n2)
- TSP: a tour of size O(n)
- BPP: an assignment (matrix) of size O(n2).
Greedy Algorithms
Key ideas:
- For many combinatorial optimization problems (but not all!), it
is easy to build a candidate solution quickly.
- Use problem structure to put together a candidate solution step-by-step.
- At each step: "do the best you can with immediate information"
- Greedy algorithms are usually O(n) or O(n2).
Example: TSP
- Greedy Algorithm:
- Initially the set P = {0, ..., n-1 } and the set
Q is empty.
- Move 0 from P to Q.
- Repeat the following until P is empty:
- Suppose k was the point most recently added to
Q.
- Find the point in P closest to k and move
that to Q.
- Output points in the order in which they were added to Q.
- What is "greedy" about this?
- At each step, we add a new point to the existing tour.
- The new point is selected based on how close it is to
previous point.
Greedy => no backtracking.
- Execution time: O(n2) (each step requires
an O(n) selection).
- Solution quality: not guaranteed to find optimal solution.
Example: BPP
- Greedy Algorithm:
- Let A = { all items }
- Sort A in decreasing order.
- At each step until A is empty:
- Remove next item in sort-order from A.
- Find first-available existing bin to fit item.
- If no existing bin can fit the item, create a new bin and
place item in new bin.
- Running time: O(n log(n)) for sort and
O(n2) for scanning bins at each step (worst-case).
=> O(n2)
- Solution quality: not guaranteed to find optimal solution.
Example: MST (Minimum Spanning Tree)
- Greedy Algorithm: (Kruskal)
- Sort edges in graph in increasing order of weight.
- Process edges in sort-order:
- If adding the edge causes a cycle, discard it.
- Otherwise, add the edge to the current tree.
- Complexity: O(E log(E)) for sorting, and O(E
log(V)) for processing the edges with union-find.
=> O(E log(E)) overall.
- Solution quality: finds optimal solution.
About greedy algorithms:
- For many problems, it's relatively easy to pose a greedy algorithm.
- For a few problems (e.g., MST), the greedy algorithm produces the
optimal solution.
- For some problems (e.g. BPP), greedy algorithms produce "reasonably
good" solutions (worst-case).
- For some problems (e.g. BPP), greedy algorithms produce "excellent"
solutions (in practice).
- For some problems (e.g., Hamiltonian tour), greedy
algorithms are of no help at all.
Computing with DNA
Yes, with actual DNA.
Key ideas:
- We will use chemical reactions with DNA to solve the
Restricted Hamiltonian Path problem
=> exploit the massive parallelism with millions of DNA molecules.
- The Restricted Hamiltonian Path problem (directed graph version):
- Given a directed graph and two vertices s and d,
find a path between s and d that visits each vertex
exactly once.
- Result: Restricted Hamiltonian Path problem is NP-complete.
- Note: for an n-vertex graph, the path is of length n.
- Overview of process:
- Represent vertices using strings of DNA bases.
- Represent edges using combinations of vertex-strings.
- Use gel-electrophoresis to extract DNA with correct length.
- Use filtering process in many steps to separate out paths
with all vertices.
=> solution (no pun intended) to Hamiltonian path problem.
Exercise:
Why is the Restricted Hamiltonian Path problem NP-complete (given that
the regular Hamiltonian Path problem is NP-complete)?
Details:
- Step 1 (on paper): associate unique DNA substrings with
vertices, e.g.
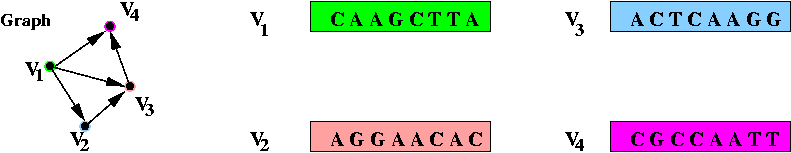
- In the above example, an 8-base string represents a vertex.
- In practice, a larger number is required to help separate out
different-length paths.
- Step 2 (on paper): associate unique DNA substrings with
edges, based on substrings for vertices:
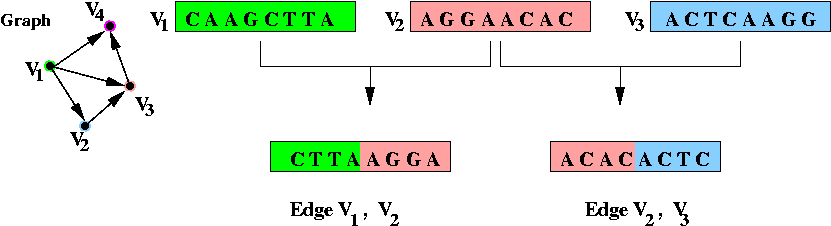
- For edge ( v1, v2) join the
latter half of v1's string with the
first half of v2's string.
- Step 3 (on paper): identify the complementary strings for
the vertices, e.g.,
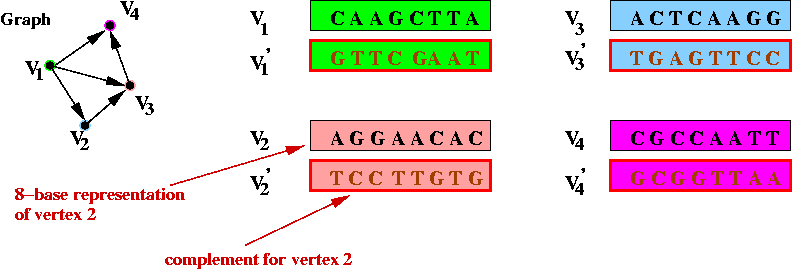
- Step 4 (on paper): create unique "start" and "stop" DNA
strings for vertices s and d.
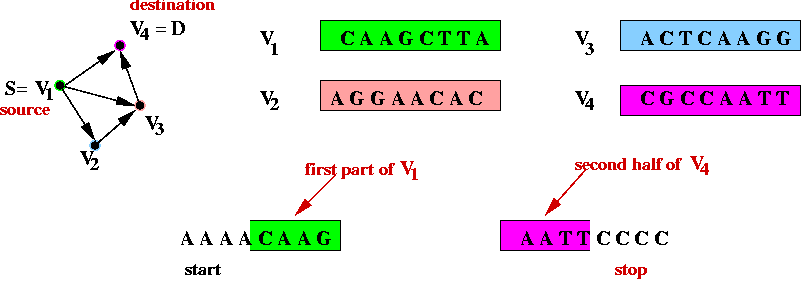
- The "start" string represents an artifical edge between
"start" and s.
- The "end" string represents an artifical edge between
d and "end".
- Step 5 (lab): synthesize all of the above DNA material
(substrings) separately (one beaker corresponding to each different string).
- Step 6 (lab): mix all the edges and complementary vertices:
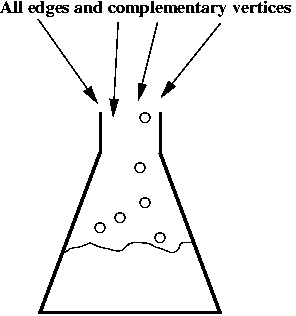
- Step 6 (lab): extract all paths beginning with "start" and
ending with "end".
=> use PCR techniques
- Step 7 (lab): separate out the DNA with the correct length
(exactly n substrings)
=> use gel-electrophoresis.
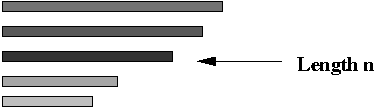
- Note: this will result in paths of exactly length n.
- However, it will also contain paths with repetitions.
- Step 8 (lab):
- Filter out all paths that don't contain the string for v1
=> use v'1 (complement) to bind.
- This leaves all paths containing v1.
- Next, filter out all paths that don't contain v2.
- ... (repeat for all vertices)
- What remains: the DNA representation of all paths of length
n that contain all vertices
=> Hamiltonian paths!
Summary:
- The purpose was to show that DNA and chemical processes can "compute".
- Potential efficiency: chemical reactions occur in parallel.
- It is not yet a practical method:
- Problems need to be carefully coded.
- Encoding takes time.
- Macro-scale experimentation results in errors
=> (fraction of a teaspoonful required for 7-vertex graph).
- Related work:
- Using DNA for building "wetware" (gates, flip-flops).
- Using proteins for computation.
- Self-assembling nanotechnology.
Genetic Algorithms and Combinatorial Optimization Problems
Key ideas:
- Use evolution as a metaphor for "winnowing" solutions.
- Outline (for TSP):
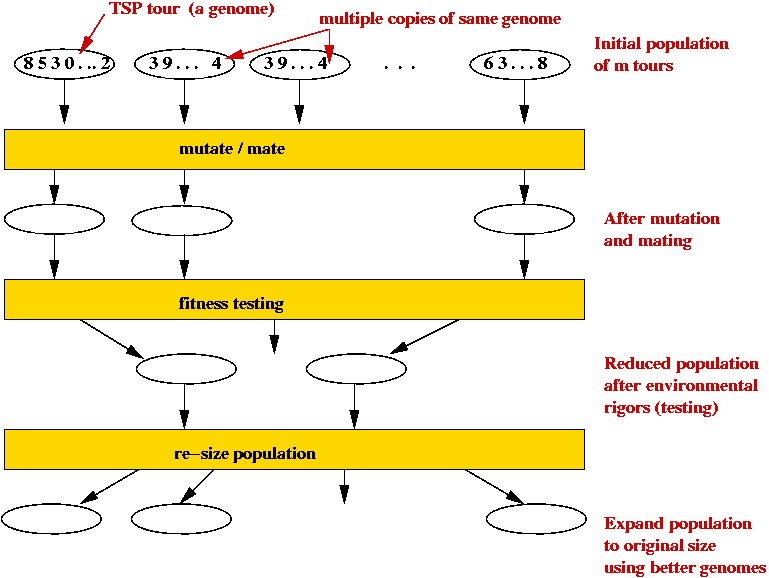
- Each candidate TSP tour is a "genome" (animal).
- Start with a large number of potential solutions (initial population).
- At each step generate a new population:
- Use mutation to "explore"
- Use mating to preserve "good characteristics".
- Weak (high-cost) solutions "die".
- Strong (low-cost) solutions "survive".
- Eventually, optimal solution should dominate population.
Details: (TSP example)
- Input: the n TSP points.
- Associate tour with genome.
- A genome's fitness is the tour's length.
(shorter the better).
- Step 1: create an initial population of m random tours
(.e.g, m = 1000).
(They don't have to be unique).
- Step 2: Compute the "fitness" value of each genome (tour).
Example with four 5-city tours:
| ID |
Genome
(tour) |
Fitness
(inverse tour length) |
Fraction of
total (PDF) |
| 1 |
0-1-2-3-4 |
27.5 |
0.31 |
| 2 |
4-0-1-3-2 |
12.95 |
0.15 |
| 3 |
0-2-1-3-4 |
9.3 |
0.11 |
| 4 |
2-4-3-0-1 |
36.0 |
0.42 |
|
|
87.75
(total) |
1.00
(total) |
- Step 3: compute the population PDF (Probability Distribution
Function)
=> fraction based on fitness.
- Compute the total fitness (sum of tour costs).
- Compute what fraction of the total each fitness value amounts to.
- The fractions are the PDF.
- Step 4: generate a new population drawing from the PDF
=> about 31% (on average) of the new population will contain
genome 1 and 11% will contain genome 3.
- Step 5: Apply crossover rules (mating):
- The crossover-fraction is an algorithm parameter,
e.g., crossover-fraction = 0.3
=> 30% of genome-pairs will engage in crossover.
- Select a random 30% of pairs randomly (assuming
crossover-fraction = 30).
- Apply a crossover rule to each such pair: exchange parts of
genomes between the pair.
- Step 6: Apply mutation
- mutation-fraction is an algorithm parameter.
- mutation-fraction = fraction of genomes to mutate
(e.g., 0.05)
- Select 5% of the genomes (randomly) to mutate.
- Apply mutation to each (make a slight adjustment in the tour).
- Repeat steps 2-6 until fitness values converge
=> population is dominated by high-fitness genomes (tours).
Crossover and mutation in TSP:
- How do we "mate" two TSP tours?
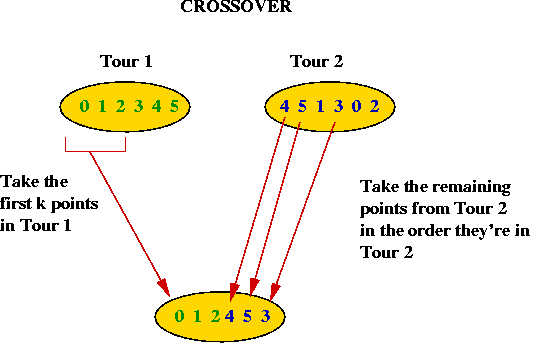
- Take the first part (about half) of Tour 1.
- Take the remaining points in the order these points are found
in Tour 2.
- How do we mutate a TSP tour?
=> swap 2 points
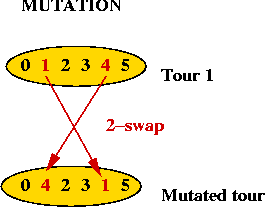
Summary:
- Advantages of genetic algorithms:
- Genetic algorithms are easy to implement.
- Like simulated annealing, the problem-specific part can be
separated out from the generic algorithm.
- If re-arrangements in fact do impact the solution, genetic
algorithms have a reasonable chance of finding a good solution.
- By its nature genetic algorithms try many initial solutions
(simultaneously)
=> simulated annealing needs to be re-run with different
starting solutions.
- Disadvantages:
- Genetic algorithms are slow.
- It's hard to define meaningful crossovers and mutations for
some problems.
- It requires some experimentation to get it working.
=> it's easier to automate this part in simulated annealing.
- Generally, simulated annealing (with appropriate
modification) is thought to be a better option.
- Warning:
- Its biological origins do not give it any special advantage
=> beware of its mystical appeal!
Neural Networks
Simplified neuron architecture:
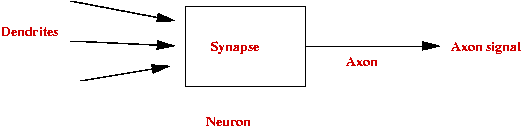
Artificial neuron:
- Structure:

- Rule: Neuron fires if X1 + X2 +
X3 > T.
A simple application to pattern recognition:
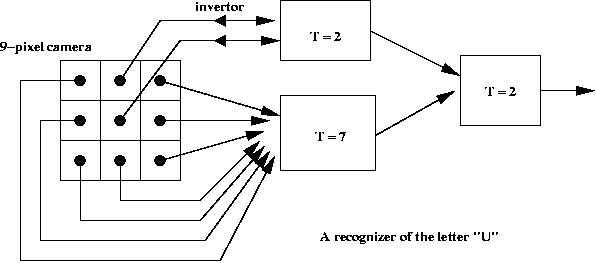
McCulloch-Pitt neuron model:
- Structure:
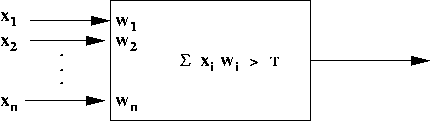
- Inputs: X1, X2, ..., Xn
- An "importance" weight Wi is associated with
each input i.
- Neuron has output only if W1X1
+ ... + WnXn > T.
Networks of neurons:
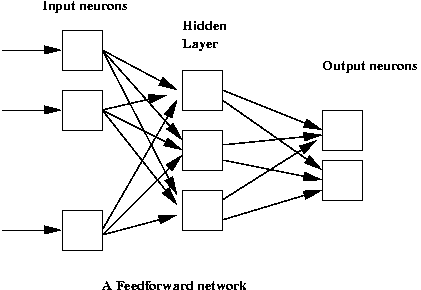
- Neural networks are used to approximate unknown functions.
- Key ideas:
- Each network has a set of parameters (neuron weights).
- Use a "training set" of samples from unknown function to set parameters.
- Once parameters are set, the function is approximated.