First, consider the search problem in genomic databases:
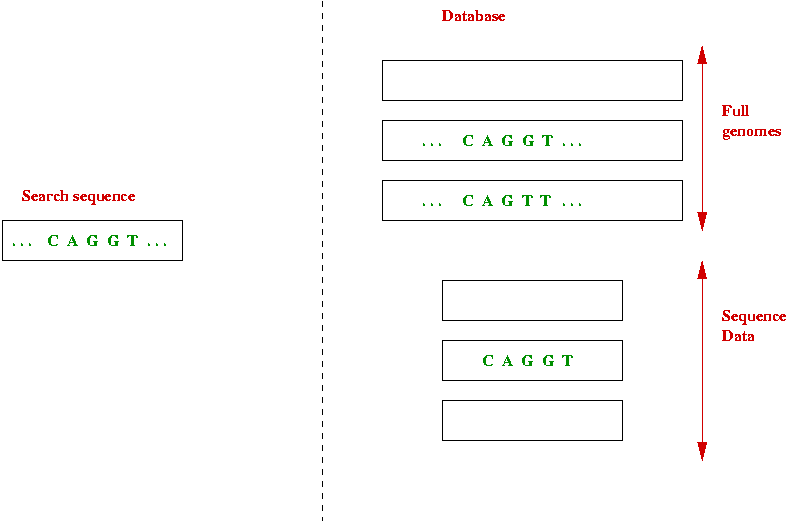
Goal: given a search sequence, find all matches in database of sequences.
Exact-match searches:
- What is an exact match?
- Easy to implement, and fast.
(See pattern
search algorithms).
- Practical use of exact-match searches: very little
- Why?
Sequence comparisons:
- We want to allow comparisons such as:
Gene comparisons:
- Recall: a gene is a substring of DNA.
- Biologists often compare two genes from different animals to
see if they are "similar"
=> to help with gene identification.
- Example: Suppose the DNA sequence A T T T C G C C G T A
C encodes a protein in animal X.
=> can search for this gene in animal Y.
- Unfortunately, genes with identical function in different
animals are not identical.
- However, they are often similar, e.g.,
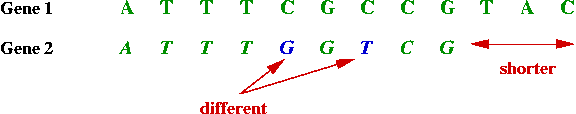
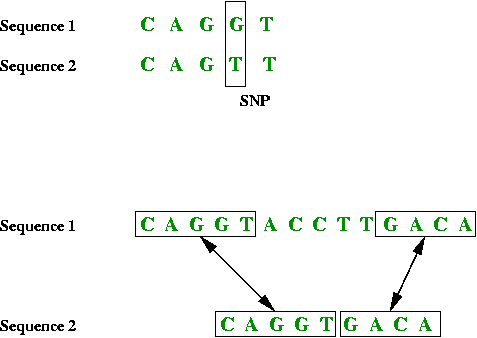
- Sometimes the similarity or "match" is more complex:
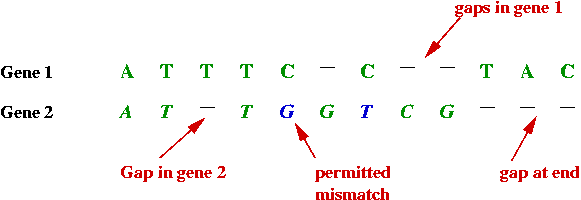
- By deliberately inserting spaces between the bases in one, a
better match can be found.
The alignment problem:
- Define scoring rules for evaluating a particular alignment:
- Perfectly matched letters get a high score.
- Matches between related letters get a modest score.
- Matches with gaps get a low score.
- The alignment problem: given two strings and the scoring
rules, find the optimal alignment.
- Problem components:
- An alphabet, e.g., { A, C, G, T }.
- Two input strings, e.g.,
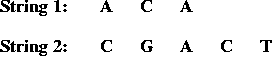
- Match and gap scores, e.g.,
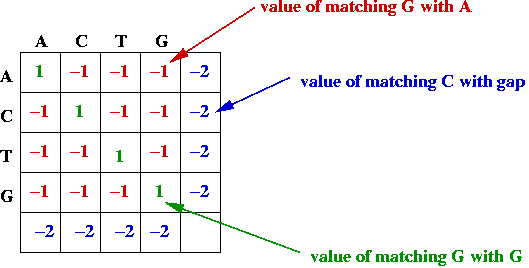
- Goal: find the alignment with the best (largest) value.
- Potential asymmetry:
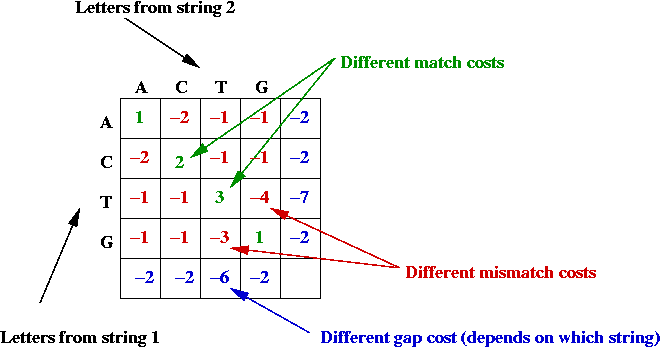
- Some letter-alignments may be more favored.
- Some gap costs are different.
- Example:
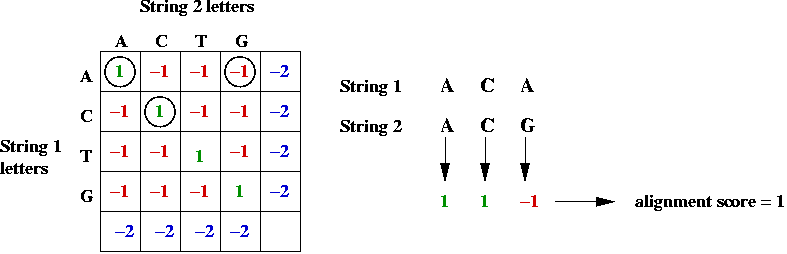
- Example:
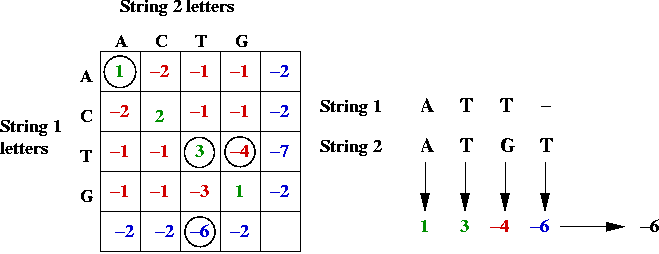
Key ideas in solution:
- First, the alignment problem can be expressed as a
graph problem:
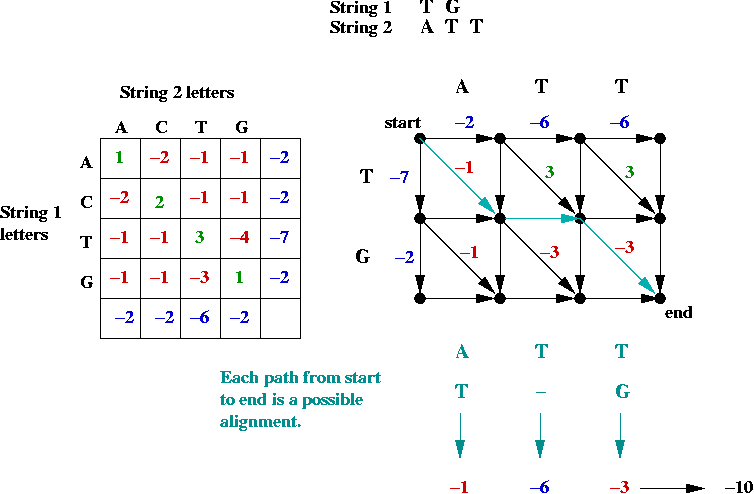
- The diagonal edges correspond to character matches
(whether identical or not).
- Each down edge corresponds to a gap in string 2.
- Each across edge corresponds to a gap in string 1.
- Goal: find the longest (max value) path from top-left to bottom-right.
=> longest-path problem in DAG.
- Can be solved in O(mn) time (using topological sort)
(m = length of string1, n = length of string2).
- Suppose we label the vertices (i, j) where
i is the row, and j is the column.
- The best path to an intermediate node is via one of its
neighbors: NORTH, WEST or NORTHWEST.
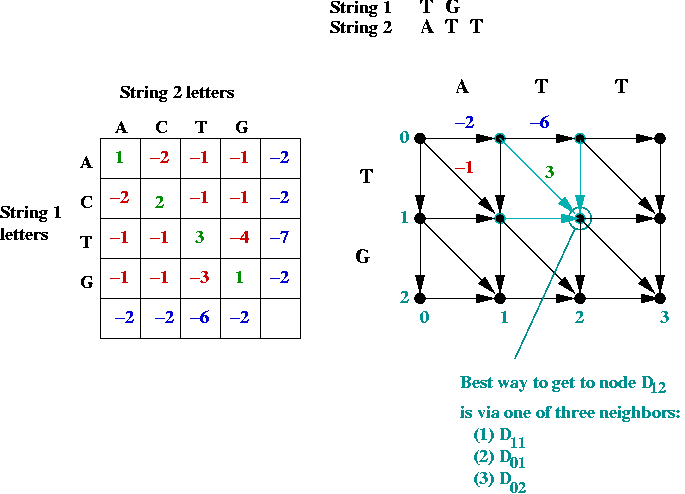
- Dynamic programming approach:
- Building an actual graph (adjacency matrix etc) is unnecessary.
- Let Di,j
= value of best path to node (i,j) from (0, 0).
= score of optimal match of first i chars in string 1
to first j chars in string 2.
- Define (for clarity)
| Ci-1,j-1 |
= |
Di-1,j-1 + exact-match-score |
| Ci-1,j |
= |
Di-1,j + string1-gap-score |
| Ci,j-1 |
= |
Di,j-1 + string2-gap-score |
- Thus,
| Di,j |
= |
max (Ci-1,j-1, Ci-1,j, Ci,j-1 |
| |
= |
max (Di-1,j-1 + exact-match-score,
Di-1,j + string1-gap-score,
Di,j-1 + string2-gap-score) |
- Initial conditions:
- D0,0 = 0
- Di,0 = Di-1,0 + gap-score for
i-th char in string 1.
- D0,j = D0,j-1 + gap-score for
j-th char in string 2.
Implementation:
- Pseudocode:
Algorithm: align (s1, s2)
Input: string s1 of length m, string s2 of length n
1. Initialize matrix D properly;
// Build the matrix D row by row.
2. for i=1 to m
3. for j=1 to n
// Initialize max to the first of the three terms (NORTH).
4. max = D[i-1][j] + gapScore2 (s2[j-1])
// See if the second term is larger (WEST).
5. if max < D[i][j-1] + gapScore1 (s1[i-1])
6. max = D[i][j-1] + gapScore1 (s2[i-1])
7. endif
// See if the third term is the largest (NORTHWEST).
8. if max < D[i-1][j-1] + matchScore (s1[i-1], s2[j-1])
9. max = D[i-1][j-1] + matchScore (s1[i-1], s2[j-1])
10. endif
11. D[i][j] = max
12. endfor
13. endfor
// Return the optimal value in bottom-right corner.
14. return D[m][n]
Output: value of optimal alignment
- Explanation:
- The 2D array D[i][j] stores the Di,j values.
- The method matchScore returns the value in matching
two characters.
- The method gapScore1 returns the value of matching a
particular character in string 1 with a gap.
(gapScore2 is similarly defined).
- Note: D[i][0] and D[0][j] represent
initial conditions.
- Note: there is one more row than the length of string 1.
(likewise for string 2).
- Computing the actual alignment:
- A separate 2D array tracks which term contributed to the
maximum for each (i, j).
- After computing all of D, traceback and obtain path (alignment).
Analysis:
- The double for-loop tells the story: O(mn) to compute Dm,n.
- Note: computing the actual alignment requires tracing back:
O(m + n) time (length of a path).
- Space required: O(mn).
An improvement (in space):
- Genes are often longer than 10,000 bases (letters) long
=> space required is prohibitive.
- First, note that we only need successive rows in computing D
=> space requirement can be reduced to O(n).
- However, O(mn) space is still required to construct
the actual alignment.
- A different approach:
- Consider the "middle" character in string 1
=> the character at position m / 2.
- In the optimal alignment, this character will either align
with some j-th character in string 1, or a gap.
=> we can try all possible j values.
- To try these alignments, compute the m/2-th row (in
O(mn) time).
- Output the optimal alignment found.
- Recurse on either side of the optimal alignment.
- It is possible to show: O(mn) time and O(n) space.
Variations of the alignment problem:
- Alignment without end-gap penalty
=> do not add costs for gaps at ends.
- Substring alignment
=> find substrings that align.
- Length-based gap penalties
=> the longer a contiguous gap, the more the penalty.
- All have polynomial-time solutions.